Results Gallery
Results from the application of pyCapsid to a set of HK97-fold protein shells.
Data covering the application of pyCapsid to this set of HK97-fold protein shells is provided here.
Prediction of disassembly units
Seneca Valley Virus (PDBID: 3cji)
pyCapsid’s b-factor predictions have a correlation coefficient of 0.64 with the experimental b-factors at 800 modes. pyCapsid identifies the rigid clusters as 12 pentons, each containing 15 MCPs. Experiments have identified that the procapsid of the virus dissociates into pentamers, matching our prediction. [1]
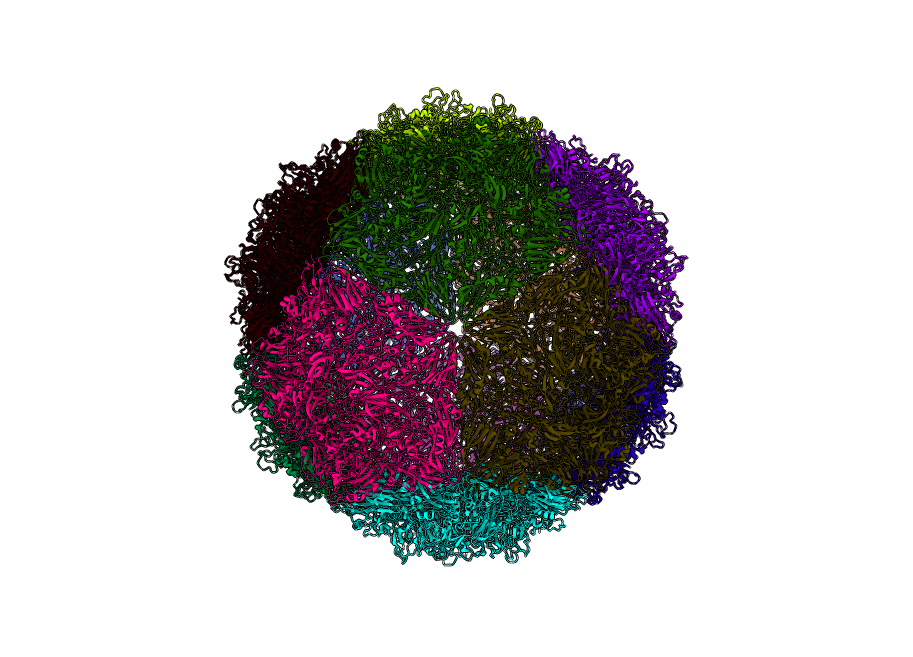
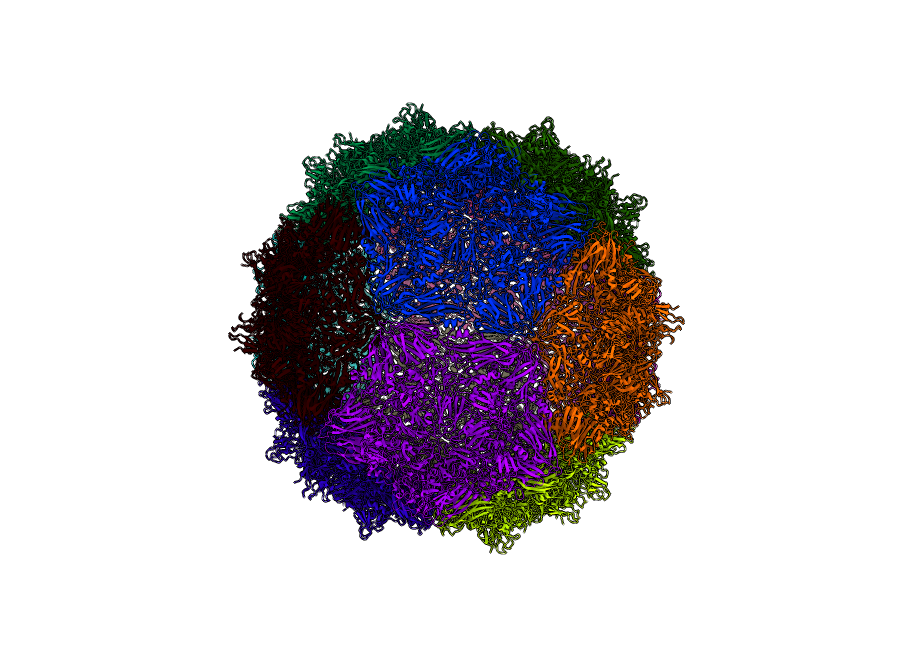
Triatoma Virus (PDBID: 3nap)
pyCapsid’s b-factor predictions have a correlation coefficient of 0.63 with the experimental b-factors at 3319 modes. pyCapsid identifies the rigid clusters as 12 pentons, each containing 15 MCPs. Both experiments and molecular dynamics simulations have been applied to explore the disassembly of this capsid, and both identify the pentons as the disassembly unit, matching our prediction. [2] [3] Both picornaviruses have similar disassembly units.
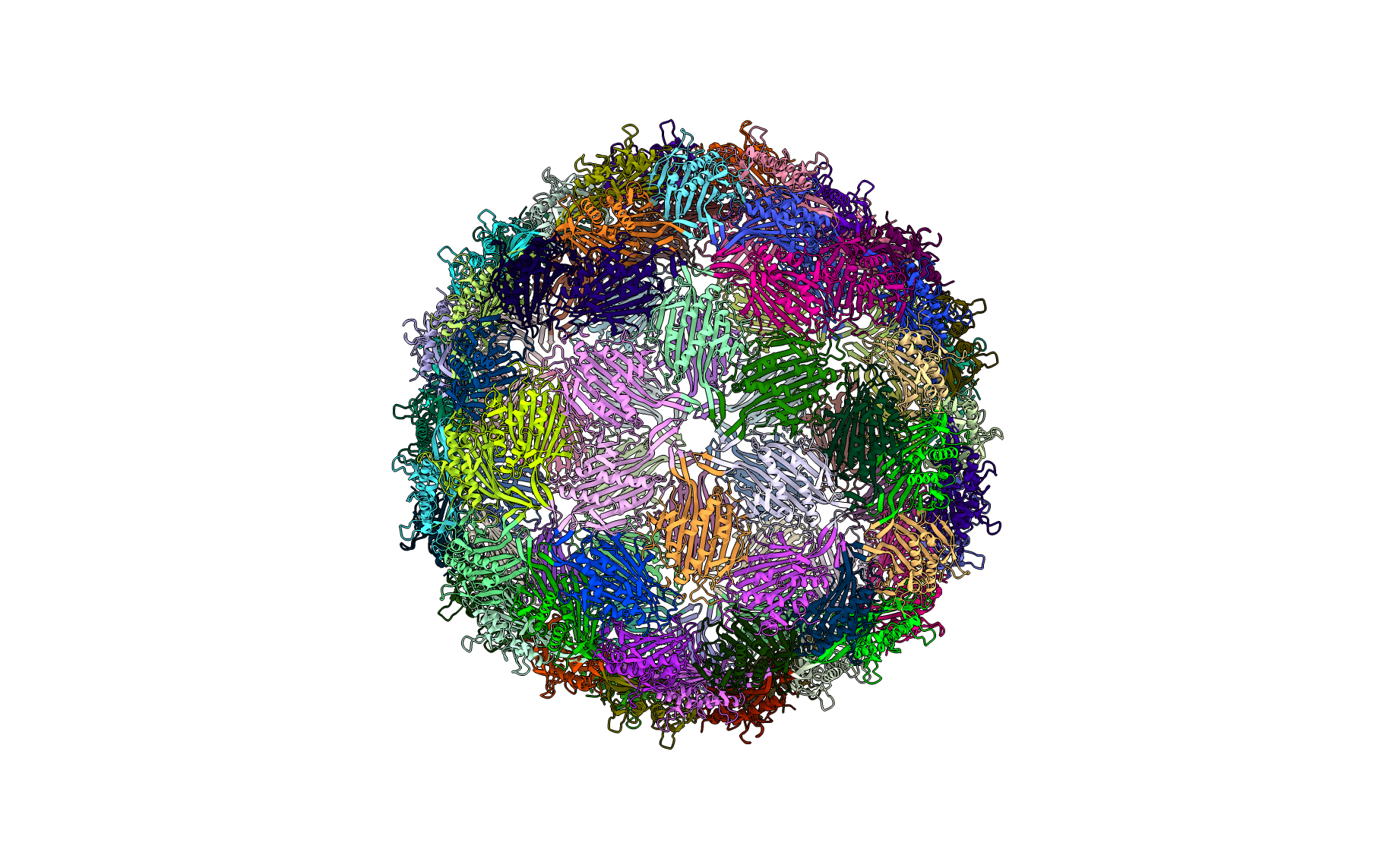
Bacteriophage MS2 (PDBID: 2ms2)
pyCapsid’s b-factor predictions have a correlation coefficient of 0.66 with the experimental b-factors at 800 modes. pyCapsid identifies the rigid clusters as 90 dimers, each containing 2 MCPs. This matches observations from NMR experiments that the capsid dissociates into dimers. [4]
Hepatitis B Virus (PDBID: 2g33)
pyCapsid’s b-factor predictions have a correlation coefficient of 0.57 with the experimental b-factors at 800 modes. pyCapsid identifies the rigid clusters as 120 dimers, each containing 2 MCPs. Both spectroscopy experiments and molecular dynamics simulations have been applied to explore the disassembly of this capsid, and both identify the dimers as the disassembly unit matching our prediction. [5] [6]
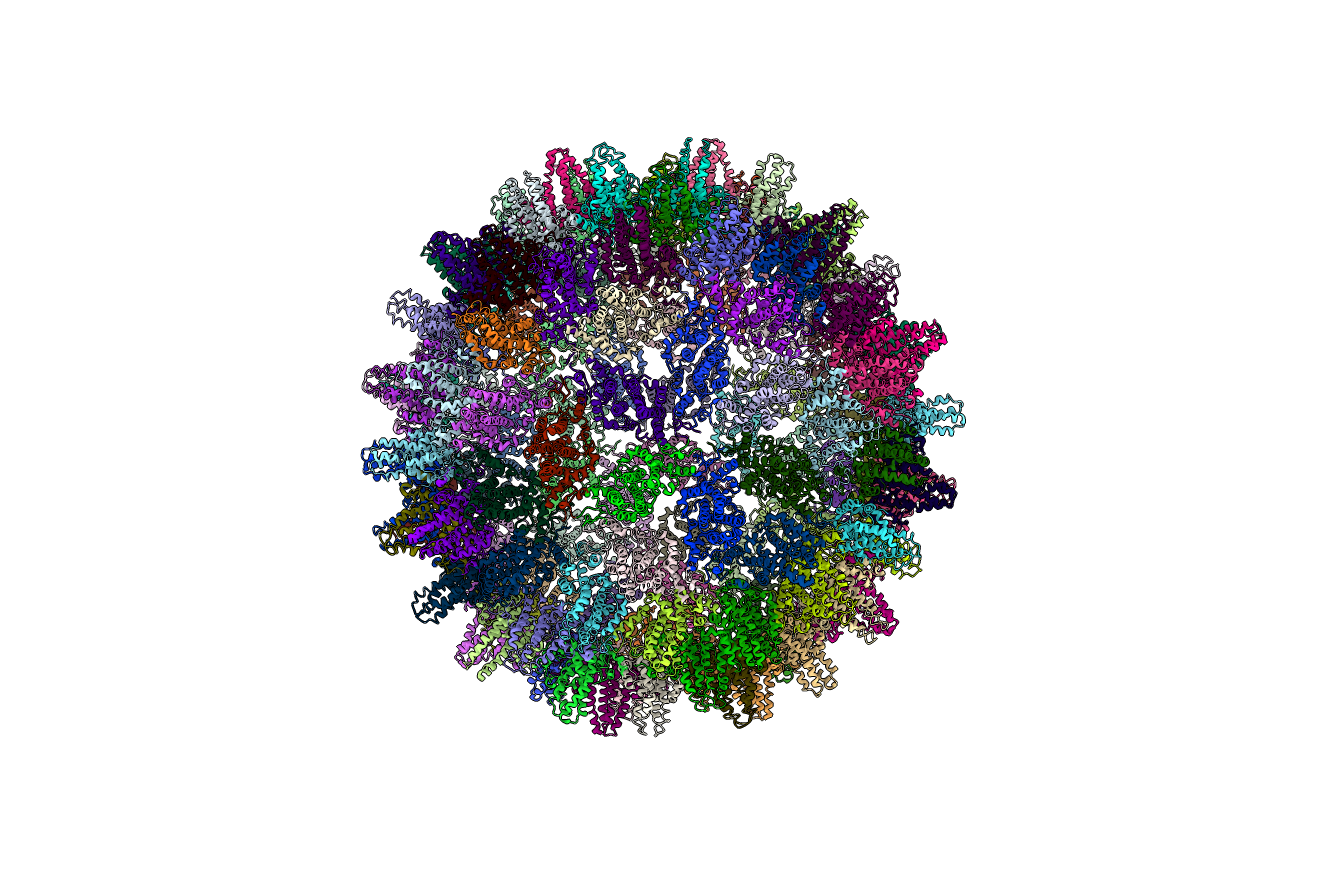
Phage P22 (PDBID: 5uu5)
pyCapsid’s b-factor predictions have a correlation coefficient of 0.6 with the experimental b-factors at 3800 modes. pyCapsid identifies the rigid clusters as either 12 pentons containing 35 MCPs each or as 12 pentamers containing 5 MCPs each and 60 hexamers containing 6 MCPs each. These correspond to the peaks in the quality score at 12 and 72. Heating the expanded shell of P22 has been shown to cause the pentamers to release, resulting in a “wiffle ball” structure. [7] This is consistent with the prediction at 72 clusters which includes pentamers as a rigid subunit.

Phage P22 Procapsid (PDBID: 2xyy)
This model has no b-factor information so results for the CC and spring constant aren’t provided. pyCapsid identifies the rigid clusters as 420 individual proteins.
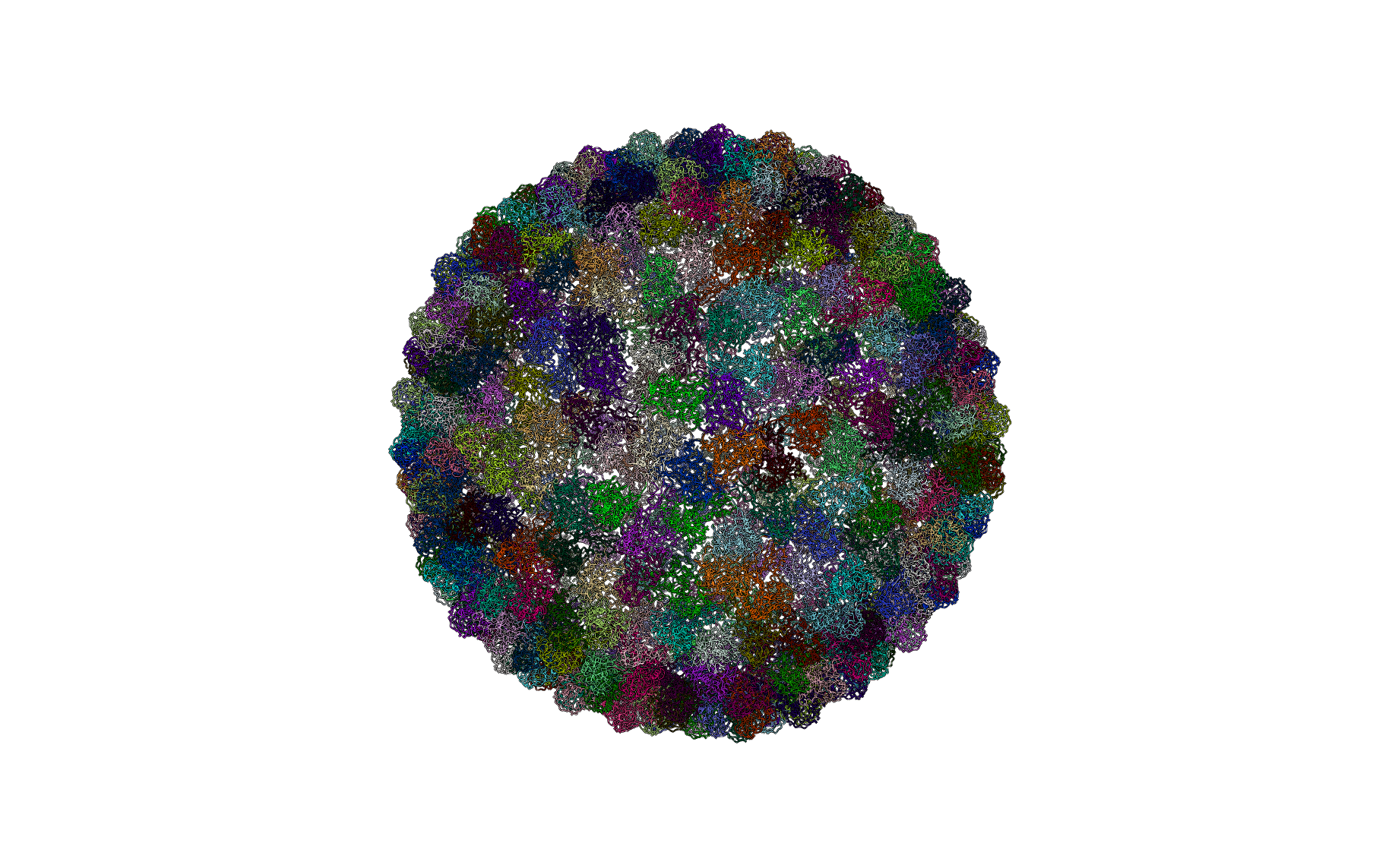
Phage HK97 (PDBID: 2ft1)
pyCapsid’s b-factor predictions have a correlation coefficient of 0.55 with the experimental b-factors at 10000 modes. pyCapsid identifies the rigid clusters as primarily the core residues of the 420 MCPs and 64 clusters corresponding to small extended domains of less than 10 residues for a total of 484 clusters. HK97 has also been observed to form the “wiffle ball” structures as in P22. [8] pyCapsid didn’t successfully predict pentameric disassembly units in this case, unlike in P22, but the prediction of individual MCPs isn’t inconsistent with pentamers.
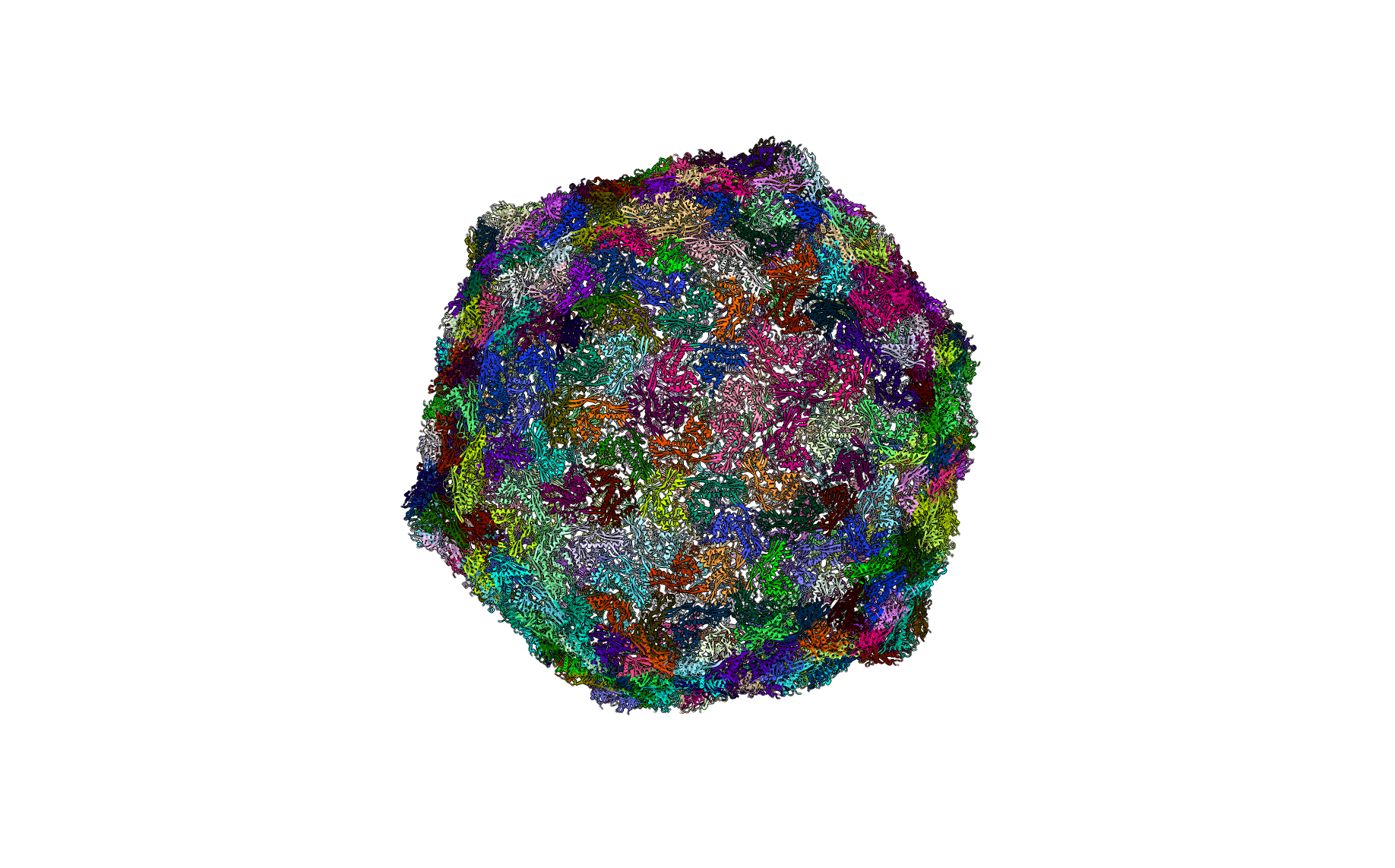
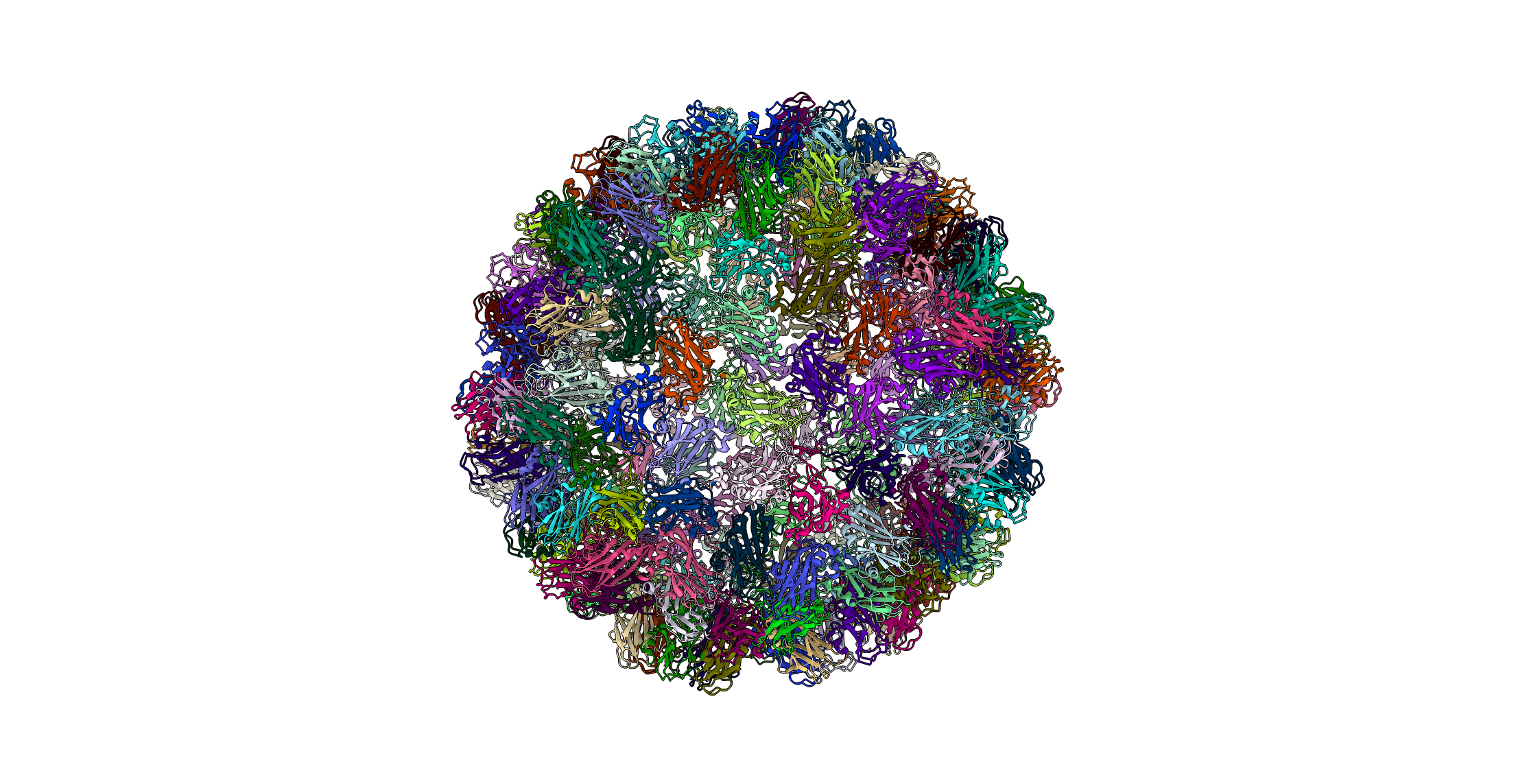
Cowpea Chlorotic Mottle Virus (PDBID: 1za7)
pyCapsid’s b-factor predictions have a correlation coefficient of 0.56 with the experimental b-factors at 6000 modes. pyCapsid identifies the rigid clusters as the 180 MCPs, one MCP per cluster. Experimental results do not support this and suggest that the capsid disassembles into dimeric groups. [9] This capsid requires a large number of modes to yield accurate correlations, with a CC of only 0.08 at 800 modes. This suggests that there may be something unique about the dynamics of the capsid that may make it more difficult for ENM/NMA methods describe. CCMV also undergoes a swelling transition before disassembly, and this intermediate swollen state may have different rigid subunits than the native state, explaning pyCapsid’s incorrect disassembly prediction. [10]
Comparisons with previous methods
PISQRD++
SPECTRUS